|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
Public Member Functions | |
| void | initialize_from_rigid (const rigid &r) |
| void | initialize_from_nrp (const non_rigid_parsed &nrp, const context &c, bool is_ligand) |
| void | initialize (const distance_type_matrix &mobility) |
Public Attributes | |
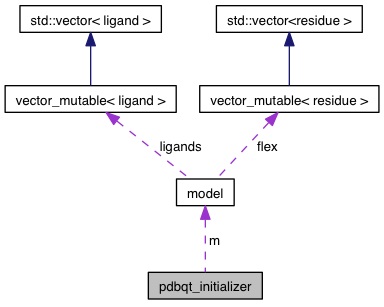
| model | m |
|
inline |
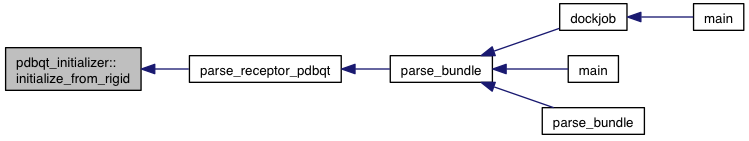
References rigid::atoms, model::grid_atoms, m, and VINA_CHECK.
Referenced by parse_receptor_pdbqt().
|
inline |
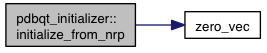
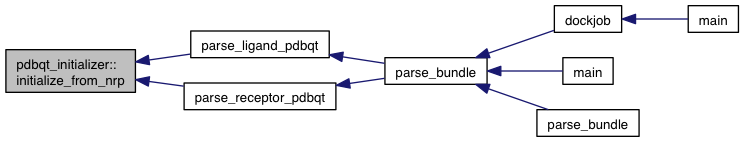
References non_rigid_parsed::atoms, model::atoms, atom::coords, model::coords, non_rigid_parsed::flex, model::flex, model::flex_context, non_rigid_parsed::inflex, model::internal_coords, non_rigid_parsed::ligands, model::ligands, m, model::m_num_movable_atoms, model::minus_forces, movable_atom::relative_coords, VINA_CHECK, VINA_FOR_IN, and zero_vec().
Referenced by parse_ligand_pdbqt(), and parse_receptor_pdbqt().
|
inline |
References model::initialize(), and m.
Referenced by parse_ligand_pdbqt(), and parse_receptor_pdbqt().
| model pdbqt_initializer::m |
Referenced by initialize(), initialize_from_nrp(), initialize_from_rigid(), parse_ligand_pdbqt(), and parse_receptor_pdbqt().