|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
#include <model.h>
Public Member Functions | |
| void | append (const model &m) |
| atom_type::t | atom_typing_used () const |
| sz | num_movable_atoms () const |
| sz | num_internal_pairs () const |
| sz | num_other_pairs () const |
| sz | num_ligands () const |
| sz | num_flex () const |
| sz | ligand_degrees_of_freedom (sz ligand_number) const |
| sz | ligand_longest_branch (sz ligand_number) const |
| sz | ligand_length (sz ligand_number) const |
| szv | get_movable_atom_types (atom_type::t atom_typing_used_) const |
| conf_size | get_size () const |
| conf | get_initial_conf () const |
| grid_dims | movable_atoms_box (fl add_to_each_dimension, fl granularity=0.375) const |
| void | write_flex (const path &name, const std::string &remark) const |
| void | write_ligand (sz ligand_number, const path &name, const std::string &remark) const |
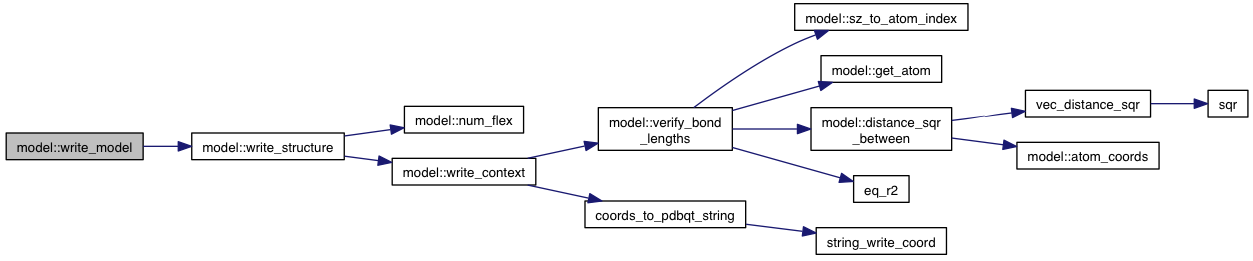
| void | write_structure (std::stringstream &out) const |
| void | write_structure (ofile &out) const |
| void | write_structure (std::stringstream &out, const std::string &remark) const |
| void | write_structure (ofile &out, const std::string &remark) const |
| void | write_model (std::stringstream &out, sz model_number, const std::string &remark) const |
| void | write_structure (const path &name) const |
| void | write_model (ofile &out, sz model_number, const std::string &remark) const |
| void | seti (const conf &c) |
| void | sete (const conf &c) |
| void | set (const conf &c) |
| fl | gyration_radius (sz ligand_number) const |
| const atom_base & | movable_atom (sz i) const |
| const vec & | movable_coords (sz i) const |
| const vec & | atom_coords (const atom_index &i) const |
| fl | distance_sqr_between (const atom_index &a, const atom_index &b) const |
| bool | atom_exists_between (const distance_type_matrix &mobility, const atom_index &a, const atom_index &b, const szv &relevant_atoms) const |
| distance_type | distance_type_between (const distance_type_matrix &mobility, const atom_index &i, const atom_index &j) const |
| fl | evali (const precalculate &p, const vec &v) const |
| fl | evale (const precalculate &p, const igrid &ig, const vec &v) const |
| fl | eval (const precalculate &p, const igrid &ig, const vec &v, const conf &c) |
| fl | eval_deriv (const precalculate &p, const igrid &ig, const vec &v, const conf &c, change &g) |
| fl | eval_intramolecular (const precalculate &p, const vec &v, const conf &c) |
| fl | eval_adjusted (const scoring_function &sf, const precalculate &p, const igrid &ig, const vec &v, const conf &c, fl intramolecular_energy) |
| fl | rmsd_lower_bound (const model &m) const |
| fl | rmsd_upper_bound (const model &m) const |
| fl | rmsd_ligands_upper_bound (const model &m) const |
| void | verify_bond_lengths () const |
| void | about () const |
| vecv | get_ligand_internal_coords () const |
| vecv | get_ligand_coords () const |
| vecv | get_heavy_atom_movable_coords () const |
| void | check_internal_pairs () const |
| void | print_stuff () const |
| fl | clash_penalty () const |
Private Member Functions | |
| model () | |
| const atom & | get_atom (const atom_index &i) const |
| atom & | get_atom (const atom_index &i) |
| void | write_context (const context &c, std::stringstream &out) const |
| void | write_context (const context &c, ofile &out) const |
| void | write_context (const context &c, ofile &out, const std::string &remark) const |
| void | write_context (const context &c, const path &name) const |
| void | write_context (const context &c, const path &name, const std::string &remark) const |
| fl | rmsd_lower_bound_asymmetric (const model &x, const model &y) const |
| atom_index | sz_to_atom_index (sz i) const |
| bool | bonded_to_HD (const atom &a) const |
| bool | bonded_to_heteroatom (const atom &a) const |
| sz | find_ligand (sz a) const |
| void | bonded_to (sz a, sz n, szv &out) const |
| szv | bonded_to (sz a, sz n) const |
| void | assign_bonds (const distance_type_matrix &mobility) |
| void | assign_types () |
| void | initialize_pairs (const distance_type_matrix &mobility) |
| void | initialize (const distance_type_matrix &mobility) |
| fl | clash_penalty_aux (const interacting_pairs &pairs) const |
Friends | |
| struct | non_cache |
| struct | naive_non_cache |
| struct | cache |
| struct | szv_grid |
| struct | terms |
| struct | conf_independent_inputs |
| struct | appender_info |
| struct | pdbqt_initializer |
| struct | model_test |
|
inlineprivate |
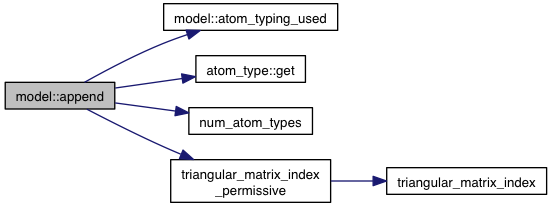
| void model::append | ( | const model & | m | ) |
References atom_typing_used(), atoms, coords, flex, flex_context, atom_type::get(), grid_atoms, internal_coords, ligands, m_num_movable_atoms, minus_forces, num_atom_types(), other_pairs, triangular_matrix_index_permissive(), VINA_CHECK, and VINA_FOR_IN.
Referenced by parse_bundle().
|
inline |
References m_atom_typing_used.
Referenced by about(), append(), eval_intramolecular(), terms::evale_robust(), initialize_pairs(), and szv_grid::szv_grid().
|
inline |
References m_num_movable_atoms.
Referenced by about(), naive_non_cache::eval(), non_cache::eval(), cache::eval(), non_cache::eval_deriv(), cache::eval_deriv(), eval_intramolecular(), terms::evale(), terms::evali(), get_heavy_atom_movable_coords(), main_procedure(), manifold_phase(), movable_atoms_box(), and non_cache::within().
| sz model::num_internal_pairs | ( | ) | const |
References ligands, and VINA_FOR_IN.
Referenced by about(), and manifold_phase().
|
inline |
References other_pairs.
Referenced by about(), and manifold_phase().
|
inline |
|
inline |
References flex.
Referenced by about(), and write_structure().
References get_branch_metrics(), branch_metrics::length, and ligands.
References branch_metrics::corner2corner, get_branch_metrics(), and ligands.
Referenced by conf_independent_inputs::conf_independent_inputs().

| szv model::get_movable_atom_types | ( | atom_type::t | atom_typing_used_ | ) | const |
References atoms, atom_type::get(), has(), m_num_movable_atoms, num_atom_types(), and VINA_FOR.
Referenced by main_procedure().
| conf_size model::get_size | ( | ) | const |
References vector_mutable< T >::count_torsions(), conf_size::flex, flex, conf_size::ligands, and ligands.
Referenced by do_search(), get_initial_conf(), main_procedure(), monte_carlo::many_runs(), monte_carlo::operator()(), manifold::operator()(), refine_structure(), and monte_carlo::single_run().
| conf model::get_initial_conf | ( | ) | const |
References get_size(), ligands, conf::ligands, conf::set_to_null(), and VINA_FOR_IN.
Referenced by do_randomization(), and do_search().

References movable_coords(), num_movable_atoms(), VINA_FOR, and VINA_FOR_IN.
Referenced by terms::evale(), and terms::evale_robust().
|
inline |
|
inline |
References flex_context, ligands, num_flex(), VINA_FOR_IN, and write_context().
Referenced by do_randomization(), write_model(), and write_structure().
|
inline |
References flex_context, ligands, num_flex(), VINA_FOR_IN, and write_context().
|
inline |
|
inline |
|
inline |
References write_structure().
Referenced by write_all_output().
|
inline |
References write_structure().
Referenced by write_structure().

| void model::seti | ( | const conf & | c | ) |
References atoms, internal_coords, ligands, conf::ligands, and vector_mutable< T >::set_conf().
Referenced by manifold_phase().


| void model::sete | ( | const conf & | c | ) |
References atoms, coords, flex, conf::flex, internal_coords, ligands, conf::ligands, vector_mutable< T >::set_conf(), and VINA_FOR_IN.
Referenced by manifold_phase().

| void model::set | ( | const conf & | c | ) |
References atoms, coords, flex, conf::flex, ligands, conf::ligands, and vector_mutable< T >::set_conf().
Referenced by do_randomization(), do_search(), monte_carlo::operator()(), manifold::operator()(), refine_structure(), and write_all_output().
References atoms, coords, EL_TYPE_H, ligands, vec_distance_sqr(), VINA_CHECK, and VINA_RANGE.
Referenced by mutate_conf().

References atoms, and m_num_movable_atoms.
References coords, and m_num_movable_atoms.
Referenced by movable_atoms_box().
| const vec & model::atom_coords | ( | const atom_index & | i | ) | const |
References coords, grid_atoms, atom_index::i, and atom_index::in_grid.
Referenced by assign_bonds(), distance_sqr_between(), and terms::evale_robust().
| fl model::distance_sqr_between | ( | const atom_index & | a, |
| const atom_index & | b | ||
| ) | const |
References atom_coords(), and vec_distance_sqr().
Referenced by assign_bonds(), atom_exists_between(), and verify_bond_lengths().
| bool model::atom_exists_between | ( | const distance_type_matrix & | mobility, |
| const atom_index & | a, | ||
| const atom_index & | b, | ||
| const szv & | relevant_atoms | ||
| ) | const |
References distance_sqr_between(), distance_type_between(), DISTANCE_VARIABLE, sz_to_atom_index(), and VINA_FOR_IN.
Referenced by assign_bonds().
| distance_type model::distance_type_between | ( | const distance_type_matrix & | mobility, |
| const atom_index & | i, | ||
| const atom_index & | j | ||
| ) | const |
References atoms, DISTANCE_FIXED, DISTANCE_VARIABLE, atom_index::i, atom_index::in_grid, and m_num_movable_atoms.
Referenced by assign_bonds(), and atom_exists_between().

| fl model::evali | ( | const precalculate & | p, |
| const vec & | v | ||
| ) | const |
References eval_interacting_pairs(), internal_coords, ligands, and VINA_FOR_IN.
Referenced by manifold_phase().

| fl model::evale | ( | const precalculate & | p, |
| const igrid & | ig, | ||
| const vec & | v | ||
| ) | const |
References coords, igrid::eval(), eval_interacting_pairs(), and other_pairs.
Referenced by eval(), and manifold_phase().
| fl model::eval | ( | const precalculate & | p, |
| const igrid & | ig, | ||
| const vec & | v, | ||
| const conf & | c | ||
| ) |
References coords, eval_interacting_pairs(), evale(), ligands, and VINA_FOR_IN.
Referenced by eval_adjusted().
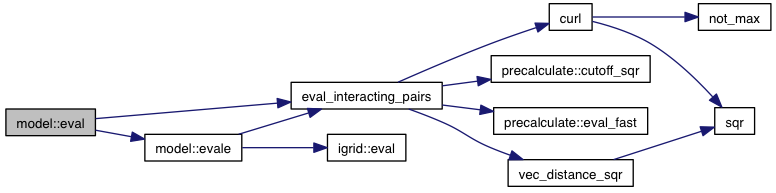
| fl model::eval_deriv | ( | const precalculate & | p, |
| const igrid & | ig, | ||
| const vec & | v, | ||
| const conf & | c, | ||
| change & | g | ||
| ) |
References coords, vector_mutable< T >::derivative(), igrid::eval_deriv(), eval_interacting_pairs_deriv(), change::flex, flex, change::ligands, ligands, minus_forces, other_pairs, and VINA_FOR_IN.
Referenced by quasi_newton_aux::operator()(), and ssd::operator()().
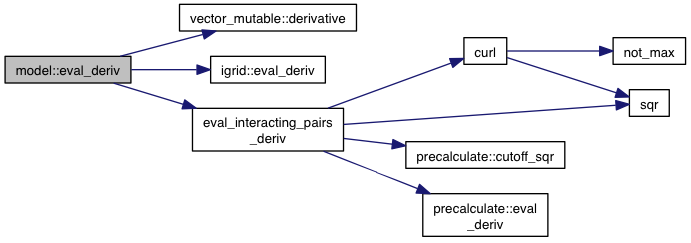
| fl model::eval_intramolecular | ( | const precalculate & | p, |
| const vec & | v, | ||
| const conf & | c | ||
| ) |
References interacting_pair::a, atom_typing_used(), atoms, interacting_pair::b, atom::coords, coords, curl(), precalculate::cutoff_sqr(), precalculate::eval_fast(), eval_interacting_pairs(), find_ligand(), atom_type::get(), grid_atoms, ligands, num_atom_types(), num_movable_atoms(), other_pairs, triangular_matrix_index_permissive(), interacting_pair::type_pair_index, vec_distance_sqr(), VINA_FOR, and VINA_FOR_IN.
Referenced by do_search().
| fl model::eval_adjusted | ( | const scoring_function & | sf, |
| const precalculate & | p, | ||
| const igrid & | ig, | ||
| const vec & | v, | ||
| const conf & | c, | ||
| fl | intramolecular_energy | ||
| ) |
References scoring_function::conf_independent(), and eval().
Referenced by do_search().
References rmsd_lower_bound_asymmetric().
Referenced by do_search().
References atom_type::ad, atoms, coords, atom_type::el, EL_TYPE_H, m_num_movable_atoms, sum(), vec_distance_sqr(), VINA_CHECK, VINA_FOR, and atom_type::xs.
Referenced by do_search().
References atom_type::ad, atoms, atom_range::begin, coords, atom_type::el, EL_TYPE_H, atom_range::end, ligands, sum(), vec_distance_sqr(), VINA_CHECK, VINA_FOR_IN, VINA_RANGE, and atom_type::xs.
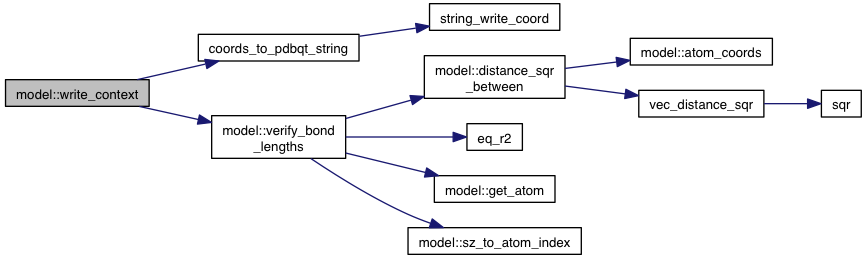
| void model::verify_bond_lengths | ( | ) | const |
References atoms, atom::bonds, bond::connected_atom_index, distance_sqr_between(), eq_r2(), get_atom(), grid_atoms, bond::length, sz_to_atom_index(), VINA_CHECK, VINA_FOR, VINA_FOR_IN, and VINA_SHOW.
Referenced by write_context().
| void model::about | ( | ) | const |
References atom_typing_used(), num_flex(), num_internal_pairs(), num_ligands(), num_movable_atoms(), num_other_pairs(), and VINA_SHOW.
Referenced by print_stuff().
|
inline |
References atom_range::begin, atom_range::end, internal_coords, ligands, VINA_CHECK, and VINA_RANGE.
|
inline |
References atom_range::begin, coords, atom_range::end, ligands, VINA_CHECK, and VINA_RANGE.
|
inline |
References atoms, coords, EL_TYPE_H, num_movable_atoms(), and VINA_FOR.
Referenced by ssd::operator()(), monte_carlo::operator()(), manifold::operator()(), and refine_structure().
| void model::check_internal_pairs | ( | ) | const |
References interacting_pair::a, interacting_pair::b, atom_range::begin, atom_range::end, ligands, ligand::pairs, VINA_CHECK, and VINA_FOR_IN.
| void model::print_stuff | ( | ) | const |
References about(), atom_type::ad, atoms, atom::bonds, atom_base::charge, atom::coords, coords, atom_type::el, grid_atoms, internal_coords, printnl(), atom_type::sy, VINA_FOR_IN, and atom_type::xs.
| fl model::clash_penalty | ( | ) | const |
References clash_penalty_aux(), ligands, other_pairs, and VINA_FOR_IN.
Referenced by do_randomization().
|
inlineprivate |
References atoms, grid_atoms, atom_index::i, and atom_index::in_grid.
Referenced by assign_bonds(), assign_types(), conf_independent_inputs::atom_rotors(), bonded_to_HD(), bonded_to_heteroatom(), terms::eval_additive_aux(), conf_independent_inputs::num_bonded_heavy_atoms(), and verify_bond_lengths().
|
inlineprivate |
References atoms, grid_atoms, atom_index::i, and atom_index::in_grid.
|
private |
References coords, coords_to_pdbqt_string(), verify_bond_lengths(), and VINA_FOR_IN.
Referenced by write_context(), write_flex(), write_ligand(), and write_structure().
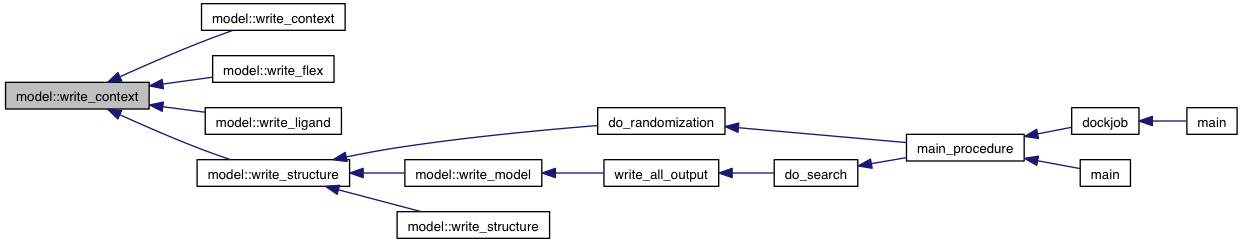
References coords, coords_to_pdbqt_string(), verify_bond_lengths(), and VINA_FOR_IN.
|
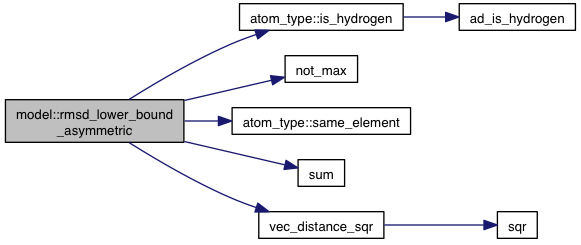
inlineprivate |
References atoms, coords, atom_type::el, EL_TYPE_H, atom_type::is_hydrogen(), m_num_movable_atoms, max_fl, not_max(), atom_type::same_element(), sum(), vec_distance_sqr(), VINA_CHECK, and VINA_FOR.
Referenced by rmsd_lower_bound().

|
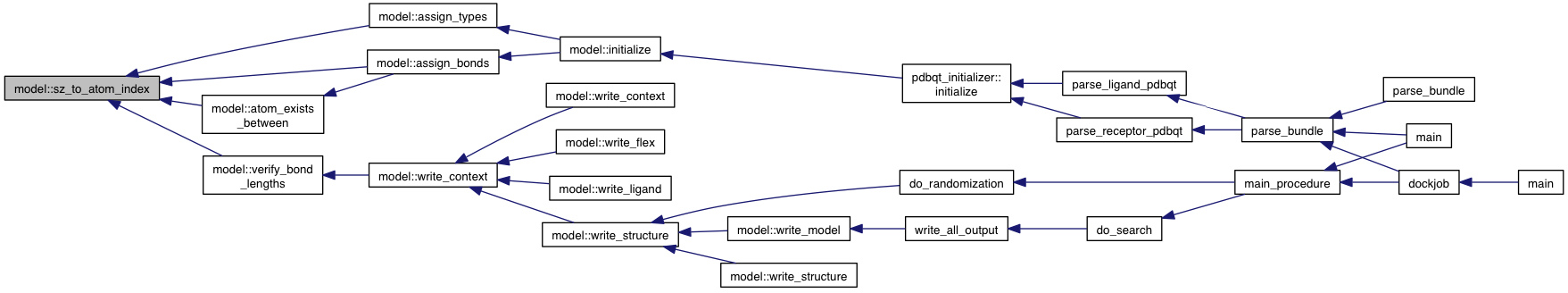
private |
References grid_atoms.
Referenced by assign_bonds(), assign_types(), atom_exists_between(), and verify_bond_lengths().
|
private |
References atom_type::ad, AD_TYPE_HD, atom::bonds, bond::connected_atom_index, get_atom(), and VINA_FOR_IN.
Referenced by assign_types().
|
private |
References atom::bonds, bond::connected_atom_index, get_atom(), atom_type::is_heteroatom(), and VINA_FOR_IN.
Referenced by assign_types().
References ligands, and VINA_FOR_IN.
Referenced by eval_intramolecular(), terms::evale_robust(), and initialize_pairs().
References atoms, bond::connected_atom_index, has(), atom_index::i, atom_index::in_grid, and VINA_FOR_IN.
Referenced by bonded_to(), and initialize_pairs().

|
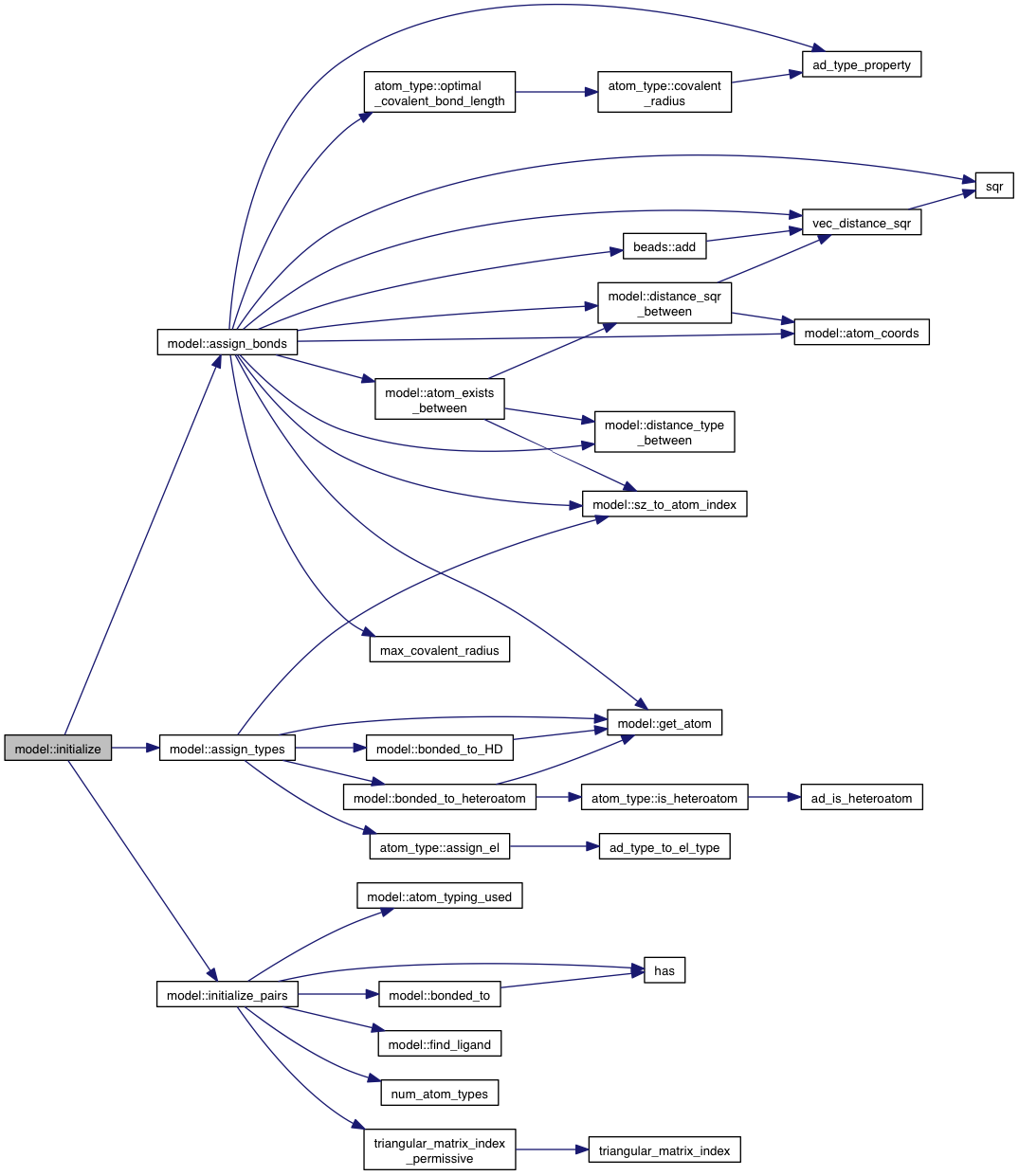
private |
References atom_type::ad, ad_type_property(), AD_TYPE_SIZE, beads::add(), atom_coords(), atom_exists_between(), atoms, atom::bonds, atom_kind::covalent_radius, beads::data, DISTANCE_ROTOR, distance_sqr_between(), distance_type_between(), DISTANCE_VARIABLE, get_atom(), grid_atoms, max_covalent_radius(), atom_type::optimal_covalent_bond_length(), sqr(), sz_to_atom_index(), vec_distance_sqr(), VINA_FOR, and VINA_FOR_IN.
Referenced by initialize().
|
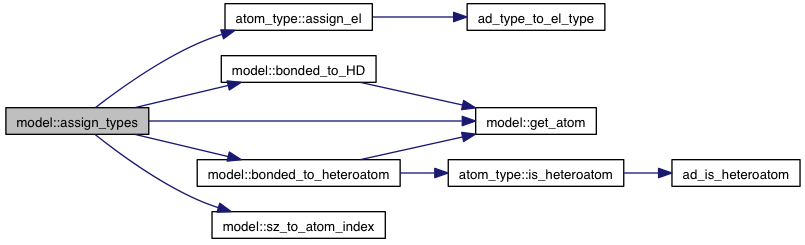
private |
References atom_type::ad, AD_TYPE_NA, AD_TYPE_OA, atom_type::assign_el(), atoms, bonded_to_HD(), bonded_to_heteroatom(), atom_type::el, EL_TYPE_Br, EL_TYPE_C, EL_TYPE_Cl, EL_TYPE_F, EL_TYPE_H, EL_TYPE_I, EL_TYPE_Met, EL_TYPE_N, EL_TYPE_O, EL_TYPE_P, EL_TYPE_S, EL_TYPE_SIZE, get_atom(), grid_atoms, sz_to_atom_index(), VINA_CHECK, VINA_FOR, atom_type::xs, XS_TYPE_Br_H, XS_TYPE_C_H, XS_TYPE_C_P, XS_TYPE_Cl_H, XS_TYPE_F_H, XS_TYPE_I_H, XS_TYPE_Met_D, XS_TYPE_N_A, XS_TYPE_N_D, XS_TYPE_N_DA, XS_TYPE_N_P, XS_TYPE_O_A, XS_TYPE_O_D, XS_TYPE_O_DA, XS_TYPE_O_P, XS_TYPE_P_P, and XS_TYPE_S_P.
Referenced by initialize().
|
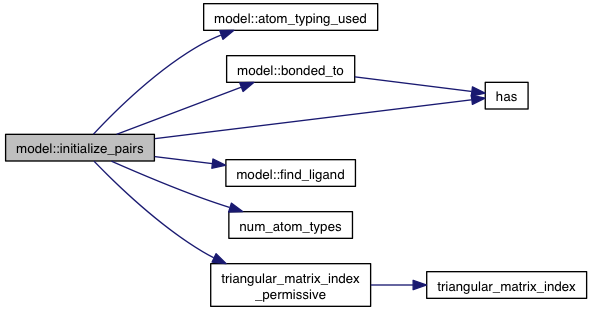
private |
References atom_typing_used(), atoms, bonded_to(), DISTANCE_VARIABLE, find_ligand(), has(), ligands, m_num_movable_atoms, num_atom_types(), other_pairs, triangular_matrix_index_permissive(), VINA_FOR_IN, and VINA_RANGE.
Referenced by initialize().
|
private |
References assign_bonds(), assign_types(), initialize_pairs(), ligands, and VINA_FOR_IN.
Referenced by pdbqt_initializer::initialize().
|
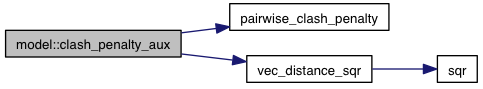
private |
References interacting_pair::a, atoms, interacting_pair::b, coords, pairwise_clash_penalty(), vec_distance_sqr(), and VINA_FOR_IN.
Referenced by clash_penalty().

|
friend |
|
friend |
|
friend |
|
friend |
|
friend |
|
friend |
|
friend |
|
friend |
|
friend |
|
private |
Referenced by append(), evali(), get_ligand_internal_coords(), pdbqt_initializer::initialize_from_nrp(), print_stuff(), sete(), and seti().
|
private |
Referenced by append(), atom_coords(), clash_penalty_aux(), naive_non_cache::eval(), non_cache::eval(), cache::eval(), eval(), non_cache::eval_deriv(), cache::eval_deriv(), eval_deriv(), eval_intramolecular(), evale(), terms::evale(), terms::evale_robust(), terms::evali(), get_heavy_atom_movable_coords(), get_ligand_coords(), gyration_radius(), pdbqt_initializer::initialize_from_nrp(), movable_coords(), print_stuff(), rmsd_ligands_upper_bound(), rmsd_lower_bound_asymmetric(), rmsd_upper_bound(), set(), sete(), non_cache::within(), and write_context().
|
private |
Referenced by append(), non_cache::eval_deriv(), cache::eval_deriv(), eval_deriv(), and pdbqt_initializer::initialize_from_nrp().
|
private |
Referenced by append(), assign_bonds(), assign_types(), atom_coords(), naive_non_cache::eval(), non_cache::eval(), non_cache::eval_deriv(), eval_intramolecular(), terms::evale(), terms::evale_robust(), get_atom(), pdbqt_initializer::initialize_from_rigid(), cache::populate(), print_stuff(), sz_to_atom_index(), szv_grid::szv_grid(), and verify_bond_lengths().
|
private |
Referenced by append(), assign_bonds(), assign_types(), bonded_to(), clash_penalty_aux(), conf_independent_inputs::conf_independent_inputs(), distance_type_between(), naive_non_cache::eval(), non_cache::eval(), cache::eval(), non_cache::eval_deriv(), cache::eval_deriv(), eval_intramolecular(), terms::evale(), terms::evale_robust(), terms::evali(), get_atom(), get_heavy_atom_movable_coords(), get_movable_atom_types(), gyration_radius(), pdbqt_initializer::initialize_from_nrp(), initialize_pairs(), movable_atom(), print_stuff(), rmsd_ligands_upper_bound(), rmsd_lower_bound_asymmetric(), rmsd_upper_bound(), set(), sete(), seti(), verify_bond_lengths(), and non_cache::within().
|
private |
Referenced by append(), check_internal_pairs(), clash_penalty(), conf_independent_inputs::conf_independent_inputs(), eval(), eval_deriv(), eval_intramolecular(), terms::evale(), terms::evale_robust(), evali(), terms::evali(), find_ligand(), get_initial_conf(), get_ligand_coords(), get_ligand_internal_coords(), get_size(), gyration_radius(), initialize(), pdbqt_initializer::initialize_from_nrp(), initialize_pairs(), ligand_degrees_of_freedom(), ligand_length(), ligand_longest_branch(), num_internal_pairs(), num_ligands(), rmsd_ligands_upper_bound(), set(), sete(), seti(), write_ligand(), and write_structure().
|
private |
Referenced by append(), eval_deriv(), terms::evale(), terms::evali(), get_size(), pdbqt_initializer::initialize_from_nrp(), num_flex(), set(), and sete().
|
private |
Referenced by append(), pdbqt_initializer::initialize_from_nrp(), write_flex(), and write_structure().
|
private |
Referenced by append(), clash_penalty(), eval_deriv(), eval_intramolecular(), evale(), initialize_pairs(), and num_other_pairs().
|
private |
|
private |
Referenced by atom_typing_used().