|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
#include <terms.h>
Public Member Functions | |
| void | add (unsigned e, distance_additive *p) |
| void | add (unsigned e, usable *p) |
| void | add (unsigned e, additive *p) |
| void | add (unsigned e, intermolecular *p) |
| void | add (unsigned e, conf_independent *p) |
| std::vector< std::string > | get_names (bool enabled_only) const |
| sz | size_internal () const |
| sz | size () const |
| sz | size_conf_independent (bool enabled_only) const |
| fl | max_r_cutoff () const |
| flv | evale (const model &m) const |
| flv | evali (const model &m) const |
| flv | evale_robust (const model &m) const |
| factors | eval (const model &m) const |
| fl | eval_conf_independent (const conf_independent_inputs &in, fl x, flv::const_iterator &it) const |
| flv | filter_external (const flv &v) const |
| flv | filter_internal (const flv &v) const |
| factors | filter (const factors &f) const |
| void | display_info () const |
Private Member Functions | |
| void | eval_additive_aux (const model &m, const atom_index &i, const atom_index &j, fl r, flv &out) const |
|
inline |
References term_set< T >::add(), and distance_additive_terms.
Referenced by everything::everything().
|
inline |
|
inline |
|
inline |
References term_set< T >::add(), and intermolecular_terms.
|
inline |
References term_set< T >::add(), and conf_independent_terms.
| std::vector< std::string > terms::get_names | ( | bool | enabled_only | ) | const |
References additive_terms, distance_additive_terms, term_set< T >::get_names(), intermolecular_terms, and usable_terms.
Referenced by current_weights(), and display_info().
| sz terms::size_internal | ( | ) | const |
References additive_terms, distance_additive_terms, term_set< T >::size(), and usable_terms.
Referenced by display_info(), eval_additive_aux(), evale(), evale_robust(), evali(), and size().
|
inline |
References intermolecular_terms, term_set< T >::size(), and size_internal().
Referenced by current_weights(), display_info(), evale(), and evale_robust().

| sz terms::size_conf_independent | ( | bool | enabled_only | ) | const |
References conf_independent_terms, term_set< T >::enabled, term_set< T >::size(), and VINA_FOR_IN.
Referenced by current_weights().


| fl terms::max_r_cutoff | ( | ) | const |
References additive_terms, distance_additive_terms, term_set< T >::max_cutoff(), and usable_terms.
Referenced by display_info(), evale(), evale_robust(), and evali().

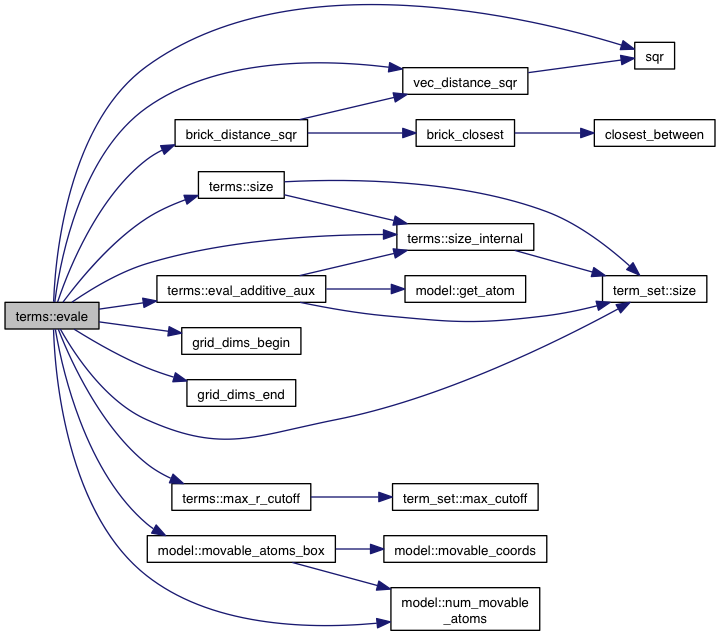
References model::atoms, brick_distance_sqr(), atom::coords, model::coords, eval_additive_aux(), model::flex, model::grid_atoms, grid_dims_begin(), grid_dims_end(), intermolecular_terms, model::ligands, max_r_cutoff(), model::movable_atoms_box(), model::num_movable_atoms(), term_set< T >::size(), size(), size_internal(), sqr(), vec_distance_sqr(), VINA_CHECK, VINA_FOR, and VINA_FOR_IN.
Referenced by eval().

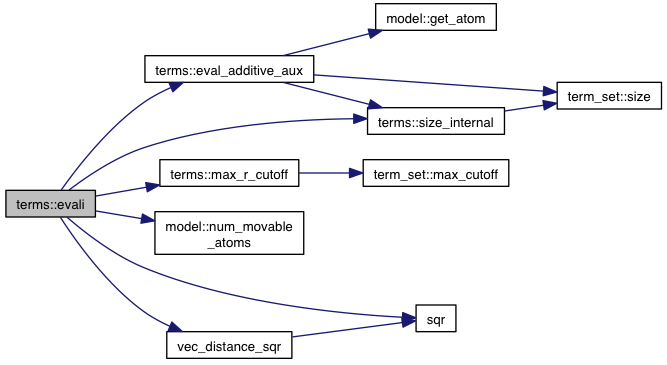
References interacting_pair::a, model::atoms, interacting_pair::b, model::coords, eval_additive_aux(), model::flex, model::ligands, max_r_cutoff(), model::num_movable_atoms(), size_internal(), sqr(), vec_distance_sqr(), VINA_CHECK, and VINA_FOR_IN.
Referenced by eval().

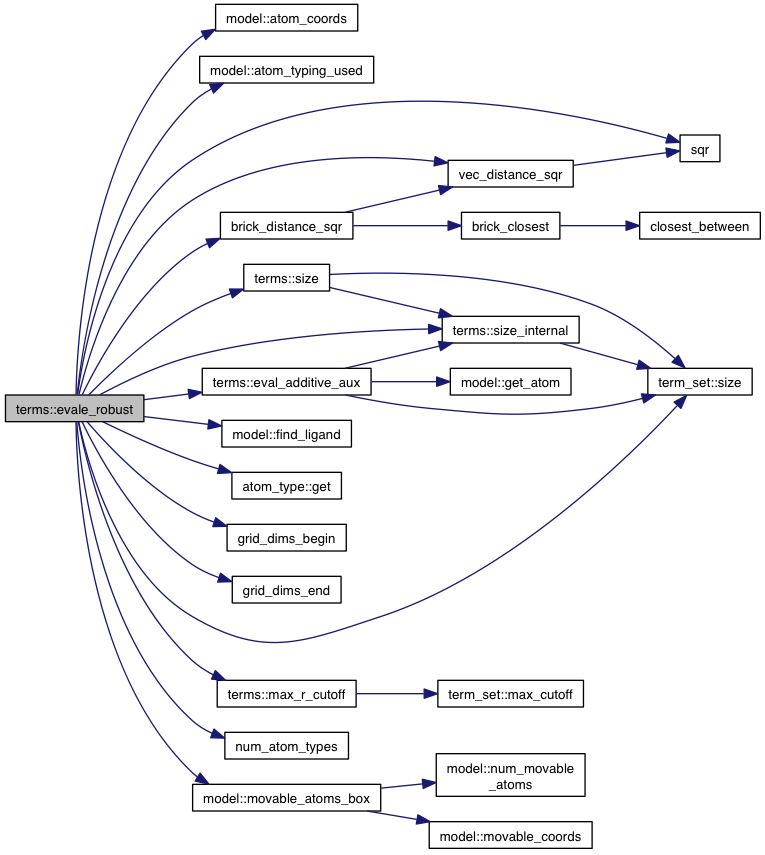
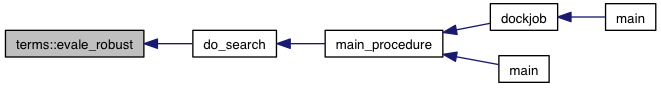
References model::atom_coords(), model::atom_typing_used(), model::atoms, atom_range::begin, brick_distance_sqr(), atom::coords, model::coords, atom_range::end, eval_additive_aux(), model::find_ligand(), atom_type::get(), model::grid_atoms, grid_dims_begin(), grid_dims_end(), intermolecular_terms, model::ligands, max_r_cutoff(), model::movable_atoms_box(), num_atom_types(), term_set< T >::size(), size(), size_internal(), sqr(), vec_distance_sqr(), VINA_CHECK, VINA_FOR_IN, and VINA_RANGE.
Referenced by do_search().
| fl terms::eval_conf_independent | ( | const conf_independent_inputs & | in, |
| fl | x, | ||
| flv::const_iterator & | it | ||
| ) | const |
References conf_independent_terms, term_set< T >::enabled, and VINA_FOR_IN.
Referenced by weighted_terms::conf_independent().
References additive_terms, distance_additive_terms, term_set< T >::filter(), intermolecular_terms, usable_terms, and VINA_CHECK.
Referenced by filter().
References additive_terms, distance_additive_terms, term_set< T >::filter(), usable_terms, and VINA_CHECK.
Referenced by filter().
References factors::e, filter_external(), filter_internal(), and factors::i.

| void terms::display_info | ( | ) | const |
References conf_independent_terms, term_set< T >::get_names(), get_names(), max_r_cutoff(), size(), size_internal(), VINA_FOR_IN, and VINA_SHOW.
|
private |
References additive_terms, distance_additive_terms, model::get_atom(), term_set< T >::size(), size_internal(), usable_terms, VINA_CHECK, and VINA_FOR_IN.
Referenced by evale(), evale_robust(), and evali().
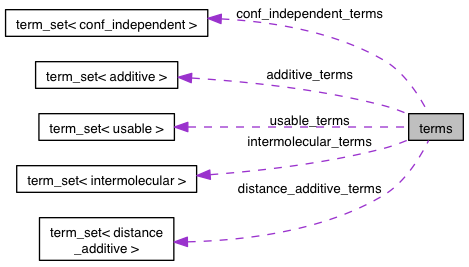
| term_set<distance_additive> terms::distance_additive_terms |
Referenced by add(), eval_additive_aux(), filter_external(), filter_internal(), get_names(), max_r_cutoff(), size_internal(), and weighted_terms::weighted_terms().
Referenced by add(), eval_additive_aux(), filter_external(), filter_internal(), get_names(), max_r_cutoff(), and size_internal().
| term_set<intermolecular> terms::intermolecular_terms |
Referenced by add(), evale(), evale_robust(), filter_external(), get_names(), and size().
| term_set<conf_independent> terms::conf_independent_terms |
Referenced by add(), display_info(), eval_conf_independent(), and size_conf_independent().