|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
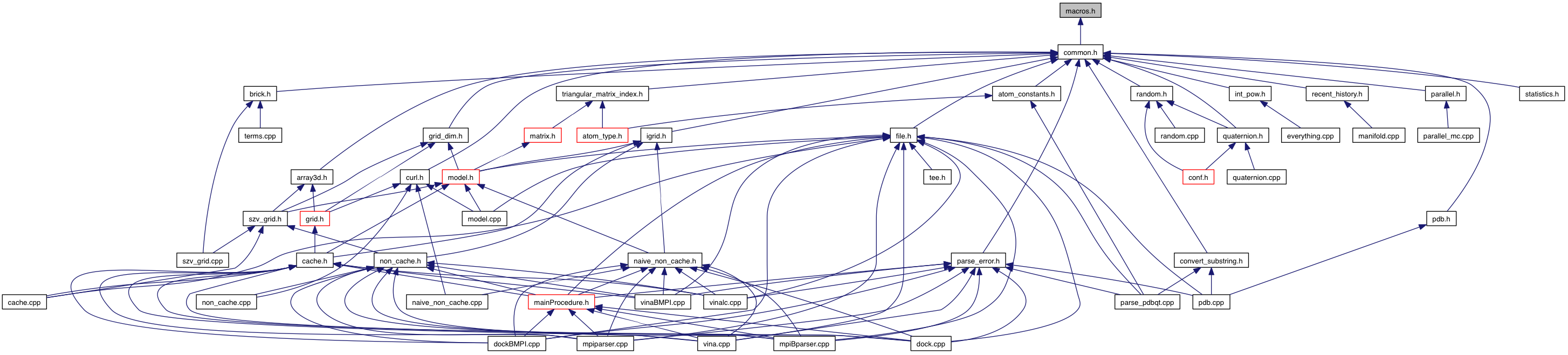
Go to the source code of this file.
Macros | |
| #define | VINA_FOR_IN(i, v) for(std::size_t VINA_MACROS_TMP = (v).size(), (i) = 0; (i) < VINA_MACROS_TMP; ++(i)) |
| #define | VINA_FOR(i, n) for(std::size_t VINA_MACROS_TMP = (n), (i) = 0; (i) < VINA_MACROS_TMP; ++(i)) |
| #define | VINA_U_FOR(i, n) for(unsigned VINA_MACROS_TMP = (n), (i) = 0; (i) < VINA_MACROS_TMP; ++(i)) |
| #define | VINA_RANGE(i, a, b) for(std::size_t VINA_MACROS_TMP = (b), (i) = (a); (i) < VINA_MACROS_TMP; ++(i)) |
| #define | VINA_U_RANGE(i, a, b) for(unsigned VINA_MACROS_TMP = (b), (i) = (a); (i) < VINA_MACROS_TMP; ++(i)) |
| #define | VINA_I_RANGE(i, a, b) for(int VINA_MACROS_TMP = (b), (i) = (a); (i) < VINA_MACROS_TMP; ++(i)) |
| #define | VINA_LOOP_CONST(t, i, v) for(t::const_iterator (i) = (v).begin(); (i) != (v).end(); ++(i)) |
| #define | VINA_LOOP(t, i, v) for(t::iterator (i) = (v).begin(); (i) != (v).end(); ++(i)) |
| #define | VINA_SHOW(x) do { std::cout << #x << " = " << (x) << std::endl; } while(false) |
| #define | VINA_SHOW_FAST(x) do { std::cout << #x << " = " << (x) << '\n'; } while(false) |
| #define | VINA_ESHOW(x) do { std::cerr << #x << " = " << (x) << '\n'; } while(false) |
| #define VINA_FOR_IN | ( | i, | |
| v | |||
| ) | for(std::size_t VINA_MACROS_TMP = (v).size(), (i) = 0; (i) < VINA_MACROS_TMP; ++(i)) |
Referenced by beads::add(), model::append(), model::assign_bonds(), model::atom_exists_between(), conf_independent_inputs::atom_rotors(), average_difference(), model::bonded_to(), model::bonded_to_HD(), model::bonded_to_heteroatom(), branches_derivative(), branches_set_conf(), brick_closest(), change::change(), pdb::check(), model::check_internal_pairs(), check_occurrence(), model::clash_penalty(), model::clash_penalty_aux(), conf::conf(), conf_independent_inputs::conf_independent_inputs(), conf_is_legal(), appender::coords_append(), count_mutable_entities(), count_torsions(), vector_mutable< ligand >::count_torsions(), vector_mutable< ligand >::derivative(), determine_iu(), deviation(), terms::display_info(), do_search(), dockjob(), eq(), parsing_struct::essentially_empty(), naive_non_cache::eval(), non_cache::eval(), weighted_terms::eval(), model::eval(), terms::eval_additive_aux(), terms::eval_conf_independent(), non_cache::eval_deriv(), model::eval_deriv(), eval_interacting_pairs(), eval_interacting_pairs_deriv(), model::eval_intramolecular(), terms::evale(), terms::evale_robust(), model::evali(), terms::evali(), conf::external_too_close(), extrapolate_cap(), term_set< additive >::filter(), find_closest(), model::find_ligand(), find_min(), conf::generate_external(), conf::generate_internal(), get_atom_range(), get_branch_metrics(), model::get_initial_conf(), term_set< additive >::get_names(), get_occurrence(), get_rankings(), grid_dims_begin(), grid_dims_end(), conf::increment(), szv_grid::index_to_coord(), model::initialize(), pdbqt_initializer::initialize_from_nrp(), model::initialize_pairs(), parsing_struct::node_t< T >::insert(), parsing_struct::node_t< T >::insert_immobiles(), parsing_struct::node_t< T >::insert_immobiles_inflex(), parsing_struct::node_t< T >::insert_inflex(), conf::internal_too_close(), main(), term_set< additive >::max_cutoff(), mean(), merge_output_containers(), precalculate_element::min_smooth_fst(), model::movable_atoms_box(), mutate_conf(), conf_independent_inputs::num_bonded_heavy_atoms(), term_set< additive >::num_enabled(), change::num_floats(), model::num_internal_pairs(), change::operator()(), parse_bundle(), cache::populate(), szv_grid::possibilities(), postprocess_branch(), postprocess_residue(), precalculate::precalculate(), change::print(), print(), conf::print(), model::print_stuff(), random_in_box(), conf::randomize(), remove_redundant(), rmsd(), model::rmsd_ligands_upper_bound(), rmsd_upper_bound(), parallel_for< aux, Sync >::run(), vector_mutable< ligand >::set_conf(), conf::set_to_null(), model::sete(), terms::size_conf_independent(), sum(), szv_grid::szv_grid(), szv_grid_dims(), torsions_generate(), torsions_increment(), torsions_randomize(), torsions_set_to_null(), torsions_too_close(), transform_ranges(), appender::update(), model::verify_bond_lengths(), weighted_terms::weighted_terms(), precalculate_element::widen_smooth_fst(), non_cache::within(), model::write_context(), and model::write_structure().
| #define VINA_FOR | ( | i, | |
| n | |||
| ) | for(std::size_t VINA_MACROS_TMP = (n), (i) = 0; (i) < VINA_MACROS_TMP; ++(i)) |
Referenced by matrix< distance_type >::append(), strictly_triangular_matrix< distance_type >::append(), appender::append(), model::assign_bonds(), model::assign_types(), szv_grid::average_num_possibilities(), bfgs_update(), precalculate::calculate_rs(), current_weights(), do_randomization(), naive_non_cache::eval(), non_cache::eval(), cache::eval(), non_cache::eval_deriv(), cache::eval_deriv(), model::eval_intramolecular(), terms::evale(), grid::evaluate_aux(), model::get_heavy_atom_movable_coords(), model::get_movable_atom_types(), grid::init(), precalculate_element::init_from_smooth_fst(), inner_product_shortest(), is_non_ad_metal_name(), manifold_phase(), monte_carlo::many_runs(), max_covalent_radius(), minus_mat_vec_product(), model::movable_atoms_box(), parallel_mc::operator()(), manifold::operator()(), mat::operator*=(), parallel_for< aux, Sync >::parallel_for(), parallel_for< F, true >::parallel_for(), pearson(), cache::populate(), precalculate::precalculate(), refine_structure(), matrix< distance_type >::resize(), model::rmsd_lower_bound_asymmetric(), model::rmsd_upper_bound(), scalar_product(), set_diagonal(), string_to_ad_type(), string_write_coord(), subtract_change(), szv_grid::szv_grid(), model::verify_bond_lengths(), precalculate::widen(), non_cache::within(), and write_all_output().
| #define VINA_U_FOR | ( | i, | |
| n | |||
| ) | for(unsigned VINA_MACROS_TMP = (n), (i) = 0; (i) < VINA_MACROS_TMP; ++(i)) |
Referenced by bfgs(), generate_external(), line_search(), manifold_phase(), ssd::operator()(), monte_carlo::operator()(), and monte_carlo::single_run().
| #define VINA_RANGE | ( | i, | |
| a, | |||
| b | |||
| ) | for(std::size_t VINA_MACROS_TMP = (b), (i) = (a); (i) < VINA_MACROS_TMP; ++(i)) |
Referenced by add_bonds(), strictly_triangular_matrix< distance_type >::append(), appender::append(), rigid_conf::apply(), bfgs_update(), pdb::check(), conf_independent_inputs::conf_independent_inputs(), terms::evale_robust(), model::get_ligand_coords(), model::get_ligand_internal_coords(), model::gyration_radius(), model::initialize_pairs(), parse_bundle(), postprocess_branch(), precalculate::precalculate(), model::rmsd_ligands_upper_bound(), atom_frame::set_coords(), substring_is_blank(), atom_frame::sum_force_and_torque(), and precalculate::widen().
| #define VINA_U_RANGE | ( | i, | |
| a, | |||
| b | |||
| ) | for(unsigned VINA_MACROS_TMP = (b), (i) = (a); (i) < VINA_MACROS_TMP; ++(i)) |
| #define VINA_I_RANGE | ( | i, | |
| a, | |||
| b | |||
| ) | for(int VINA_MACROS_TMP = (b), (i) = (a); (i) < VINA_MACROS_TMP; ++(i)) |
| #define VINA_LOOP_CONST | ( | t, | |
| i, | |||
| v | |||
| ) | for(t::const_iterator (i) = (v).begin(); (i) != (v).end(); ++(i)) |
| #define VINA_LOOP | ( | t, | |
| i, | |||
| v | |||
| ) | for(t::iterator (i) = (v).begin(); (i) != (v).end(); ++(i)) |
| #define VINA_SHOW | ( | x | ) | do { std::cout << #x << " = " << (x) << std::endl; } while(false) |
Referenced by model::about(), terms::display_info(), and model::verify_bond_lengths().
| #define VINA_SHOW_FAST | ( | x | ) | do { std::cout << #x << " = " << (x) << '\n'; } while(false) |
| #define VINA_ESHOW | ( | x | ) | do { std::cerr << #x << " = " << (x) << '\n'; } while(false) |