|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
#include <szv_grid.h>
Public Member Functions | |
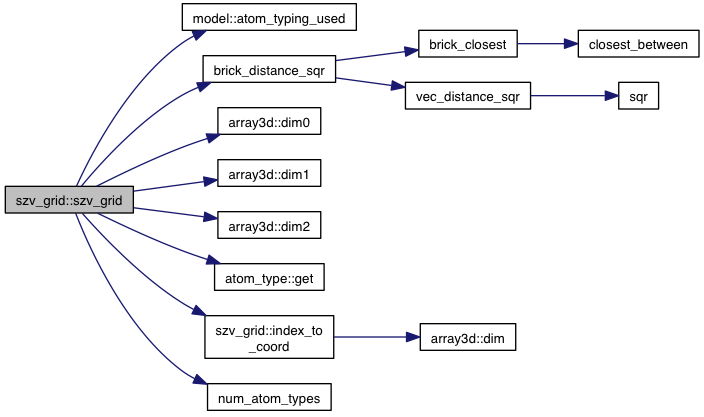
| szv_grid (const model &m, const grid_dims &gd, fl cutoff_sqr) | |
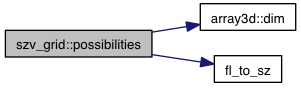
| const szv & | possibilities (const vec &coords) const |
| fl | average_num_possibilities () const |
Private Member Functions | |
| vec | index_to_coord (sz i, sz j, sz k) const |
Private Attributes | |
| array3d< szv > | m_data |
| vec | m_init |
| vec | m_range |
References model::atom_typing_used(), brick_distance_sqr(), atom::coords, array3d< T >::dim0(), array3d< T >::dim1(), array3d< T >::dim2(), atom_type::get(), model::grid_atoms, index_to_coord(), m_data, m_init, m_range, num_atom_types(), VINA_FOR, and VINA_FOR_IN.
References array3d< T >::dim(), fl_to_sz(), m_data, m_init, m_range, and VINA_FOR_IN.
Referenced by non_cache::eval(), non_cache::eval_deriv(), and cache::populate().
| fl szv_grid::average_num_possibilities | ( | ) | const |
References array3d< T >::dim0(), array3d< T >::dim1(), array3d< T >::dim2(), m_data, and VINA_FOR.
References array3d< T >::dim(), m_data, m_init, m_range, and VINA_FOR_IN.
Referenced by szv_grid().
Referenced by average_num_possibilities(), index_to_coord(), possibilities(), and szv_grid().
|
private |
Referenced by index_to_coord(), possibilities(), and szv_grid().
|
private |
Referenced by index_to_coord(), possibilities(), and szv_grid().