|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
|
|
VinaLC: Parallel Molecular Docking Program |
Biochemical and Biophysical Systems Group |
#include <non_cache.h>
Public Member Functions | |
| non_cache (const model &m, const grid_dims &gd_, const precalculate *p_, fl slope_) | |
| virtual fl | eval (const model &m, fl v) const |
| virtual fl | eval_deriv (model &m, fl v) const |
| bool | within (const model &m, fl margin=0.0001) const |
Public Attributes | |
| fl | slope |
Private Attributes | |
| szv_grid | sgrid |
| grid_dims | gd |
| const precalculate * | p |
| non_cache::non_cache | ( | const model & | m, |
| const grid_dims & | gd_, | ||
| const precalculate * | p_, | ||
| fl | slope_ | ||
| ) |
Implements igrid.
References precalculate::atom_typing_used(), model::atoms, atom::coords, model::coords, curl(), precalculate::cutoff_sqr(), precalculate::eval_fast(), gd, atom_type::get(), get_type_pair_index(), model::grid_atoms, num_atom_types(), model::num_movable_atoms(), p, szv_grid::possibilities(), sgrid, slope, sqr(), VINA_FOR, and VINA_FOR_IN.
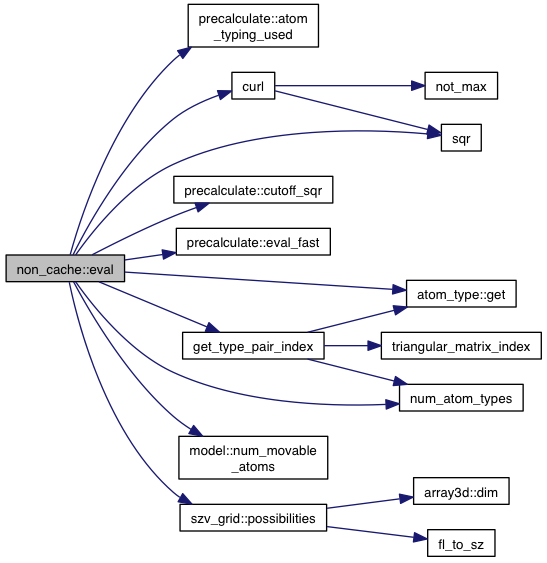
Implements igrid.
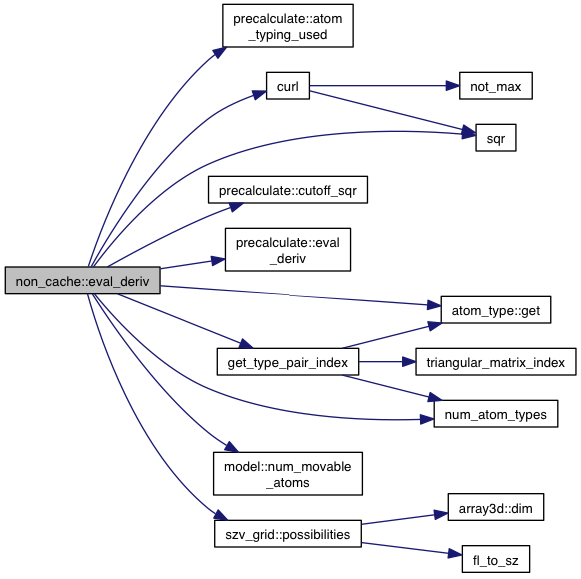
References precalculate::atom_typing_used(), model::atoms, atom::coords, model::coords, curl(), precalculate::cutoff_sqr(), precalculate::eval_deriv(), gd, atom_type::get(), get_type_pair_index(), model::grid_atoms, model::minus_forces, num_atom_types(), model::num_movable_atoms(), p, szv_grid::possibilities(), sgrid, slope, sqr(), VINA_FOR, and VINA_FOR_IN.
References model::atoms, model::coords, gd, model::num_movable_atoms(), VINA_FOR, and VINA_FOR_IN.
Referenced by do_search(), and refine_structure().
| fl non_cache::slope |
Referenced by eval(), eval_deriv(), and refine_structure().
|
private |
Referenced by eval(), and eval_deriv().
|
private |
Referenced by eval(), eval_deriv(), and within().
|
private |
Referenced by eval(), and eval_deriv().